Humans in general feel a need to create order by organizing the things in their environment – hands up those who keep their t-shirts and underwear in separate drawers, and/or arrange them in order of most recently worn or colour. Coupled with the universal - some teenagers excluded - desire for order is a related compulsion to name things. The great Chinese thinker and philosopher K'ung Fu Tzu (better known by his Latin name, Confucius) is widely credited as the source of the old Chinese proverb: “The beginning of wisdom is to call things by their proper name.” But, what’s the point of naming things? Why do we bother?
We name objects because it makes our lives easier. Let’s say you’re sitting on the sofa and you want your friend to pass the remote so you can see what else is on the TV – this process is rather difficult without names. A request like, “Please pass the thing on the thingy. I want to see what’s on the whats-am-a-jig”, is likely to be met with confusion. The request is easier for the other person to follow if things have names: “Please pass the remote on the coffee table. I want to see what’s on the TV”. Now, it’s true that you might be able to gesticulate at your friend until he or she either gets the idea, or misinterprets and takes offence; but what if you can’t see the person you need help from – charades doesn’t help then.

Imagine that you’re sitting on the train going to work when you remember you forgot to get the pie out of the freezer to defrost in time for dinner. Fortunately, your partner has the day off and is at home, so you phone up and ask “Can you get the thing out of the thingy so it’s thingy-ed in time for what’s-its-name?” Again, confusion reigns. Gesticulating won’t help because the other person can’t see you, although it might make a dull train ride more interesting for your fellow passengers. The instructions can be followed when we insert the names: “Can you get the pie out of the freezer so that it’s defrosted in time for dinner?”
So, the act of naming facilitates communication. Whether the objects are pieces or furniture, bits of machinery, or animals, we assign them names because it makes life easier for us. We, for example, call a ‘fish’ with a cartilaginous skeleton and between five and seven pairs of gills a “shark”. This allows us to tell another person what animal we’re looking at or talking about. The use of a name certainly helps, but is not without problems. Telling someone that you went diving with sharks while on holiday can be like saying you had meat for dinner; it’s not quite as specific as we might want because there are lots of different ‘types’ of meat (and sharks). Consequently, to make our meaning as clear as possible, to designate with accuracy and precision, objects (be they animals, plants, bacteria, furniture, tools, etc.) are split into as narrow groups as realistically possible and each group is given a name.
So, for example, the group of ‘fish’ we call sharks is further subdivided into different types of sharks based largely on how they look (their “morphology”), both internally (i.e. their skeleton, internal organs etc.) and externally (i.e. fins, gills, skin, colour etc.). Large groups are then split into smaller, more specific ones and so on down the line until you have a group containing all the animals considered to be exactly the same in terms of the features (morphological, genetic, ecological, biochemical, even behavioural) under consideration. This is the species level, and we’ll look at this in more detail later. Humans, chimpanzees, great white sharks, blackbirds, palmate newts and red squirrels are all examples of species.
Some taxonomists opt to take the splitting below the species level and group animals into subspecies, infraspecies and forms, among others. Perhaps the extreme of this splitting is found in the human species, where every individual of the species is given his or her own name at birth. The problem with being so specific, however, is that it quickly gets complicated as the list of viable names runs out, forcing several individuals to share the same name – think how confusing it can be if there are two or three people in the office with the same name. Consequently, the branch of Science known as Taxonomy, from the Greek word taxis meaning “order” or “rank” and –nomia, meaning “law”, is largely concerned with the grouping of organisms down to the species level.

This system of giving each species a name is all well and good and it certainly makes it easier to be precise in our communications - but, in order for the system to work, everyone must call that “something” by the same universally agreed name – if the process isn’t regulated we can run into problems. Such problems are rife with “common names”. Here in the UK, we have a stunning bird of prey called the peregrine falcon; the fastest bird species in the world, clocked at speeds of 87mph / 140kmph during a dive. In North America, however, the same bird is more commonly known as the duck hawk, after its impressive ability to nab ducks in mid-air. Anyone who wasn’t aware of this “double identity” could reasonably assume that we were talking about two different species. The problem gets exponentially more complicated when local names, different languages and different dialects are considered.
So, how do we get around this? Well, we do it by giving most species known to Science two names: a vernacular (common) and a scientific (often referred to as Latin, but more accurately a Latinized-Greek) one. While it’s true that not all species have a vernacular name (e.g. many bacteria, mosses, lichens etc.), this isn’t a major issue because it is the Latin name that’s the important one; it’s designed to remove confusion caused by dual identities.
I will return to our falcon example shortly, but first let’s take a brief look at how we arrived at the classification system we embrace today and how we use it to assign animals a unique Latin name.

Birth of the binomen
Carl von Linné (also variously referred to as Carl Linnaeus, Carolus Linnaeus and, more colloquially, the "Father of Taxonomy"), is largely responsible for the way we classify creatures today. Linné was born in Sweden during May 1707, and transferred from a study of medicine to a study of plants in 1728. In 1735, he returned to his study of medicine, completing it before going on to publish the first edition of his classification of living things, titled Systema Naturae, in which he listed all types of animals that he knew of. Systema Naturae began life as a small pamphlet, but by 1758, when the tenth edition was published, it had become a multi-volume opus. Not only did Linnaeus list all the animals he knew about; he also grouped them by his own hierarchical scheme according to how closely related he thought they were, which he based on how similar they looked to one another.
Despite some controversial aspects, Linnaeus’ scheme has proven to be robust and much of it remains to this day. The system comprises a series of levels, or categories, called taxa (singular being taxon) and assigns each species a binominal name. All scientific names ascribed to species are initially binomial (i.e. they are composed of two parts), consisting of a generic (i.e. genus-related) and a specific (i.e. species-related) epithet. Where the controversial process of further splitting occurs, the organism may be assigned a trinomial (three-part) name, to show that it’s a subspecies. So, for example, in the case of the red fox the binomial name is Vulpes vulpes, but some genetic data suggest the populations in Europe and North America are sufficiently different to be classed at least as separate subspecies; those in Europe are Vulpes vulpes vulpes (the originally described population is given the species name as the subspecies and we call this the nominate subspecies), while those in North America are Vulpes vulpes fulva.
In the standard taxonomic hierarchy, there are seven taxa, with the species name sitting at the lowest, or “basic”, level. In other words, ignoring subspecies for now, the species name represents the narrowest grouping. While seven taxa are pretty standard in a classification scheme, the total number can be substantially higher; the largest I’ve seen was 76. The number of taxa, and the names ascribed to them, can also vary according to whether the species you’re classifying is an animal, plant, bacteria etc. Nonetheless, regardless of the number of taxa sitting above it, the species level is the only one that can truly be considered “natural”, because everything above it is largely subjective – different taxonomists may place a given species in different taxa, but the species should remain the same.
One for all and all for one

Each taxonomist generally has his or her own ideas about how animals and plants are related to each other, and few ever agree on a single (universal) taxonomic scheme for anything. Arguably, this doesn’t really matter; these fanciful Latin/Greek names are constructs of our own convenience and have no relevance in the wider natural world. After all, whether I file my ELO CD under Rock or Pop doesn’t change the CD itself, any more than choosing to classify a duck within the Tubulinida (the class containing the amoebas) makes it less of a bird and more of a protozoa in real life. The scheme we use just represents how closely related we think the critter in question is to other critters assigned to different species. As a result, no single taxonomic scheme is inherently "better" than another. All that really matters is that the resulting scheme best fits the available evidence.
Having read this far, you might be wondering what the point of classifying animals is if it has little relevance in nature. Well, classification is essential when it comes to drawing up protection for species. In a fascinating article in Scientific American, published in June 2008, science writer Carl Zimmer provided a nice example of this involving wolves. In the southern USA, there is a considerable conservation effort to save the red wolf, which is considered a separate species from the wolves in Canada and the eastern USA. Some scientists, however, argue that the red wolf is just an isolated population of the Canadian species, which -- if true -- means that the US government hasn’t actually been saving a species from extinction, because thousands still reside just across the border. In his article Zimmer also noted that proper classification of microbes could allow public health workers to anticipate outbreaks of disease and prepare a response.
So, the point here is that taxonomy is about more than just scientists arguing over which scheme they think best suits a given species; it has deep roots in our understanding of species relationships and in the protection of the natural world. Anyway, enough with the preamble – how do we actually put plants, animals and microbes into these groups?
Taxonomic levels
I have already mentioned that there are at least 76 taxonomic levels that can be used to build a detailed classification. Seven taxa are, however, usually sufficient, and they are: Kingdom; Phylum (from the Greek phulon, meaning “race”); Class; Order; Family; Genus; and Species. Precisely how these are defined and allocated varies according to the type of organism (animal, plant, bacteria etc.) you’re trying to classify.

In Linnaeus’ original scheme, objects were grouped into one of three Kingdoms: Animalia (animals); Vegetabilia (plants); or Mineralia (minerals) – hence the familiar “animal, vegetable or mineral” options. As our knowledge of the natural world grew, indeed keeps growing, taxonomists found that these three kingdoms weren’t sufficient to do justice to the enormous diversity of life on Earth. We now recognize six kingdoms: Plantae (plants); Animalia (animals); Fungi (fungi and moulds); Eubacteria (the bacteria, sometimes called Monera); Archaea (microbes similar to bacteria); and the Protista (a dumping ground for multi-cellular organisms that don’t fit into any of the aforementioned groups, sometimes called Protoctista). Despite some quite apparent differences between the two, a few textbooks merge the Eubacteria and Archaea into a single kingdom: the Prokaryota.
Depending on the scheme you choose to follow (and they change frequently), the kingdoms break down roughly as follows:
- Plantae is divided into about 12 phyla and comprise about 270,000 species.
- Animalia is split into about 33 phyla and contains about 800,000 species (although this is probably a drastic underestimate of the true figure).
- Fungi have five phyla and about 100,000 species.
- Eubacteria have three phyla and a number of species that is difficult even to estimate – some authors suggest 1,000,000,000 (a billion) but even this could be a considerable underestimate.
- Archaea are poorly known and there are currently three main (and five tentative) phyla that have been created based largely on laboratory cultures (estimates of total phyla range from 18 to 23). The most recent list I can find (1999) contains 209 species.
- Protista comprise some 20 to 50 phyla and about 23,000+ species.
If we dig a little deeper and look at an example of a ‘standard’ classification, we can see how these taxa are arranged. In the structure below I have set out the currently accepted taxonomic scheme for that most infamous of all sharks, the great white.
Kingdom: Animalia (mobile critters; have many cells; can’t make their own food)
Phylum: Chordata (flexible skeletal rod with accompanying nerves)
Class: Chondrichthyes (‘fish’ with a cartilaginous skeleton)
Order: Lamniformes (‘Mackerel’ sharks)
Family: Lamnidae (‘Mackerel’ sharks)
Genus: Carcharodon (from the Greek carcharos meaning “ragged” or “pointed” and odon meaning “tooth”)
Species: carcharias (Greek for “shark”)
Working down from the above, the scheme moves from a very broad taxon (i.e. Animalia with thousands of species), to a slightly narrower one (the class containing just the cartilaginous fishes with almost 1,500 species), to a narrower one still (containing only ‘mackerel’ sharks, about 15 species) and so on down to the narrowest one (i.e. species – just one).
Origin of a scientific name
The scientific name given to an organism is usually based either on a description of it, the region in which it’s found, or the person describing it for the first time; sometimes a combination of these. The name Myotis macrotarsus, for example, was given to a cave-roosting bat from the Philippines and translates roughly to “mouse-eared bat with big feet” – an apt description of the critter! Similarly, in our scheme above, the genus and species epithet combine to form a rather pertinent description of the great white shark (pointed-toothed shark). The giant otter, on the other hand, was first described from Brazil and is given the scientific name Pteronura brasiliensis, while the South African lantern shark, Etmopterus compagnoi, was named after taxonomist Professor Leonard Compagno at the South African Museum.
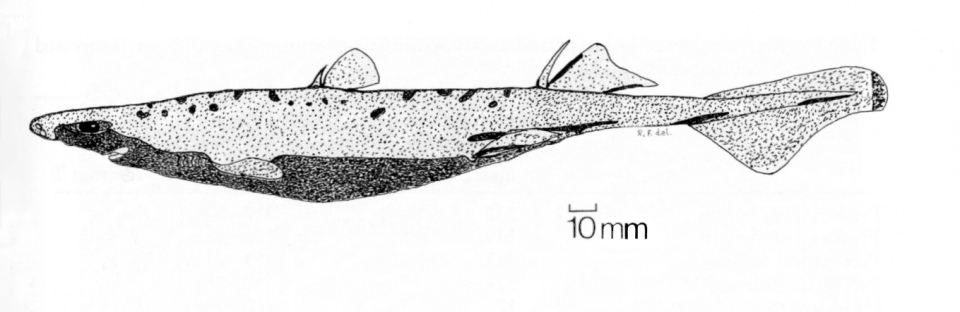
In a few instances, an organism may be given a scientific name that illustrates a particular behaviour – a good example of this can be found in fish called scats. Scats are allocated the genus Scatophagus, from the Greek skatos meaning “dung” or “faeces” and phagein meaning “to eat”, after their penchant for eating monkey excrement that falls into the water.
We have looked at several examples of how the scientific names have Latin/ancient Greek origins. It should be mentioned, however, that all taxa names -- not just species names -- have their roots in Greek or Latin and can also be roughly translated into English. Returning to our white shark scheme, for example, the order Lamniformes can be broken down into Lamni- (from the Greek lamna meaning “voracious fish”) and –formes (from the Latin forma, meaning “shape”) so that the sharks in this order are all of “voracious fish shape”.
At this point, you might be wondering why we bother with Latin/Greek names? Why not just use English, or any other of the 6,909 globally recognized languages? Latin was once widely used among Renaissance scholars throughout what is now Europe, allowing people in one country to effectively communicate with someone else in another country, much like English today. Latin and ancient Greek, however, are both considered ‘dead’ languages, which means that they’re no longer learnt as a native tongue and are thus no longer evolving. To put it another way, the Latin word forma means “shape” today and meant “shape” a century ago – as you can imagine, this is not the case for many of the languages in use today, especially English.
Above the species level, most taxa have standardized formal and informal suffixes, which helps to clarify their position. For example, almost all families are formed by adding the ending -idae (animals) or -aceae (plants) to the stem of the genus name. So, the main genus in the dog family is Canis and it sits within the family we formally call the Canidae. The informal suffix for families is usually –id. This means we can say members of the Canidae family are canids. Similar rules work for subfamilies (-ine and -inane), but it is rather more complicated for orders.
Speaking and writing

Latin names can appear daunting, especially when it comes to trying to pronounce them. For example, take Acrocephalus schoenobaenus (the Latin name of a bird called the sedge warbler), Plectrophenax nivalis (the snow bunting, another small bird) or Mertensiella luschani (a salamander found in the Aegean). The longest currently-accepted Latin name goes to the Indian soldierfly Parastratiosphecomyia stratiosphecomyioides. How would you go about pronouncing any of these?
The best advice is to break down and ‘sound out’ the words. So, for example, our sedge warbler would be broken down something like: Acro-ceph-alus scho-en-o-bae-nus. In Latin, most of the vowels are short; also, ch is pronounced as a “k”, ae as “ee”, and ph as “f”. So, if you ‘sound out’ our example, it would be: Acro-cef-a-lus skoo-en-o-bee-nus. A little practice and you’ll soon pick it up! One final point to remember is that not everyone agrees exactly how Latin or ancient Greek words should be pronounced (George Hempl wrote about this at some length in 1898), so don’t be surprised if you hear others ‘correcting’ you, or pronouncing them differently – don’t take it personally!
When it comes to writing Latin names, there are a couple of rules that should be followed. The first is that the genus is always capitalized (i.e. begins with a capital letter), while the specific name is not. So, in the case of our sedge warbler, the Latin name should always be Acrocephalus schoenobaenus and never Acrocephalus Schoenobaenus or acrocephalus schoenobaenus. Also, the scientific name should be italicized wherever possible and underlined where italics are not available, such as in handwritten documents. Finally, only the genus and species epithets should be italicized/underlined – the kingdom, phylum, order, family etc., should not be in italics, despite having the same Greek/Latin origin.
Regulation of scientific names
The ultimate goal of binomial nomenclature -- nomenclature being a set, or system, of names or terms -- is to remove the confusion that vernacular (common) names sometimes cause. Remember back to our example of the peregrine falcon, known in the USA as the duck hawk. Despite having two common names, it only has one Latin name: Falco peregrinus (falco is Latin for “hawk”, while peregrinus is Latin for “wandering”). If you were to write “I saw a peregrine (Falco peregrinus) today” it should leave people, both in the UK and in the USA, in no doubt which bird you’re talking about. So, with this in mind, it becomes apparent that Latin names only work if each species has one, and only one, binomen. This is indeed the case and no two species can have the same scientific name – or, more specifically, the same species epithet.
The task of governing the system for ensuring that every animal has a unique and universally accepted scientific (binomial) name falls to the International Commission on Zoological Nomenclature (ICZN). Founded in 1895, the ICZN now has 28 members spread across 20 countries and sees some 2,000 new generic and 15,000 new specific names added to or restored in the zoological literature each year. The ICZN has the final say on whether a proposed scientific name should be uniformly accepted by the zoological community. Opinions of the ICZN are published in their own quarterly journal, the Bulletin of Zoological Nomenclature.
The scientific names of all plants and fungi are regulated by two primary codes: The International Code of Botanical Names and The International Code of Nomenclature of Cultivated Plants, published by the International Botanical Congress (IBC). The naming of bacteria is mitigated under the International Code of Nomenclature of Bacteria, published by the International Committee on Systematics of Prokaryotes (ICSP). The classification of viruses is currently slightly different to other groups, but is overseen by the International Committee on Taxonomy of Viruses (ICTV).
Overall, in order for a species to be accepted as distinct from any other, a formal description of it must be published in the scientific literature and a “type” (representative) specimen must be preserved in a museum or university so it can be used as a standard by which to compare other specimens. When considering which names to attribute to a species, it is the oldest valid (published) name that has priority – it is the overseeing authority’s (i.e. ICZN, IBC, ICTV or ICSP) job to settle any nomenclatorial disputes.
Cladistics: Ancestry, shared features and the task of classification
The object of any good biological taxonomic system is that it represents what we currently know of the evolutionary relationships of its subjects. Most taxonomic schemes arrange organisms in terms of the shared characteristics that they possess: probably the most popular way of doing this is with cladistics (from the Greek klados, meaning “branch” or “rank”). The basic objective of cladistics is to provide a scheme showing the most likely evolutionary pathway for a given group or species based on the characters that it shares with its relatives.
The premise behind cladistics is delightfully simple: if the feature that you’re looking at is present in two different organisms then it is likely to have been inherited from their most recent common ancestor. That said, as the late elasmobranch biologist Aidan Martin noted in his article on the subject: not all features are equally useful when looking at ancestry. Features that abound among different organisms are retained because they suit a purpose, even though their owners may since have diverged from the common ancestor (Mr. Martin referred to these as “evolutionary hangers on”). In the article, Aidan wrote:
“… a two-opening gut (with a mouth at one end and a cloaca or anus at the other) is an ancestral character. Both you and a cockroach have a two-opening gut, but you would probably take offence if I were to suggest that you and a cockroach were closely related …”
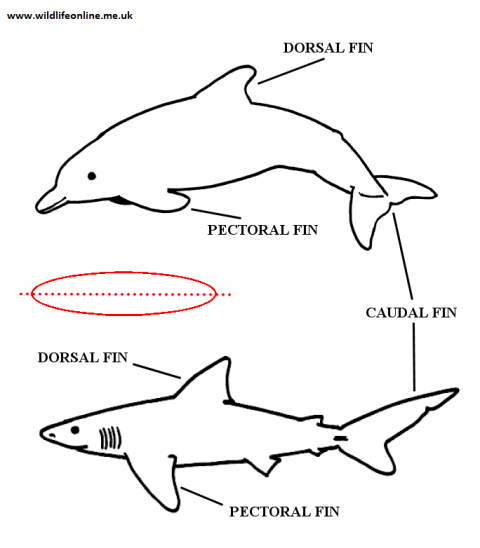
In effect, with cladistics we are looking for modifications of long-running characteristics - variations on a theme, if you like. Consequently, in order to undertake a cladistic analysis we must translate whatever it is we observe into discrete characters. The ability to translate traits into discrete units (i.e. present or absent with no in-betweens) means that cladistics lends itself well to computer analysis.
The language of taxonomy can be a little confusing and I will gloss over most of the terminology as it doesn't concern us here. There are, nonetheless, a few 'central terms' that crop up a lot. When it comes to looking at traits, there are two main types: homoplasic and homologous. Homoplasic (not to be confused with homoplastic) traits are those that bear no relationship to the relatedness of two individuals – they have remained because they suit the environment in which their owner lives. So, for example, sharks and dolphins share a similar body form -- i.e. fusiform (torpedo-shaped) body, with similar-looking fin arrangements -- because this is best suited to an aquatic lifestyle; they're not closely related. We call this convergent evolution. When taxonomists use the term "homology", however, they're talking about a similarity of traits in two or more species (or groups) that's the result of them sharing a common ancestor at some time in the past. When thinking about homologies, there are two basic character states: plesiomorphic and apomorphic (or "derived").
When you’re comparing two organisms, they will invariably exhibit characters that are shared widely with other groups or species (these are the plesiomorphic, or “ancestral” traits) and others that are unique to them or their group (these are the apomorphic/derived traits). It is sometimes said that plesiomorphic/ancestral characters are “primitive”, while apomorphic/derived traits are “advanced” – most taxonomists shy away from these terms, though, because they’re easily misconstrued. So, a trait that is present in lots of different species or groups, such as the twin-opening gut, is plesiomorphic and doesn’t give us any clues as to our species’ ancestry.
Conversely, those features that are present only in an ancestor and its descendants are apomorphic and can be used to assess taxonomic relationships. Characters that are unique to a species (i.e. have arisen within the species and aren’t present in any ancestors) are referred to as autapomorphic. It is important to recognise that all these terms are relative; a character can be an apomorphy at one branch of your tree, but plesiomorphic at another. Feathers, for example, characterize (i.e. are apomorphic for) the group we call Aves (birds), but is plesiomorphic to peregrine falcons. In other words, having feathers can be used to define an animal as a member of the Aves, but not to define it as a peregrine falcon because all birds have feathers and they’re not taxonomically unique to this species.
When a character is present in two or more species and originated in their most recent common ancestor, the feature is called a synapomorphy. Finally, a character shared by several groups or species having originated in a distant ancestor (i.e. older than the most recent common one) is referred to as symplesiomorphic. When you have a group that, based on synapomorphies, contains the common ancestor and all species descended from it, you have what taxonomists refer to as a monophyletic (meaning “one race”) group – these are also sometimes referred to as “natural groups” or “clades”. The opposite of this, where you have a group that contains an ancestor and some of its descendants, is a paraphyletic (“near race”) group. A third option is a polyphyletic group, which is based on homoplasy and doesn’t contain a common ancestor.
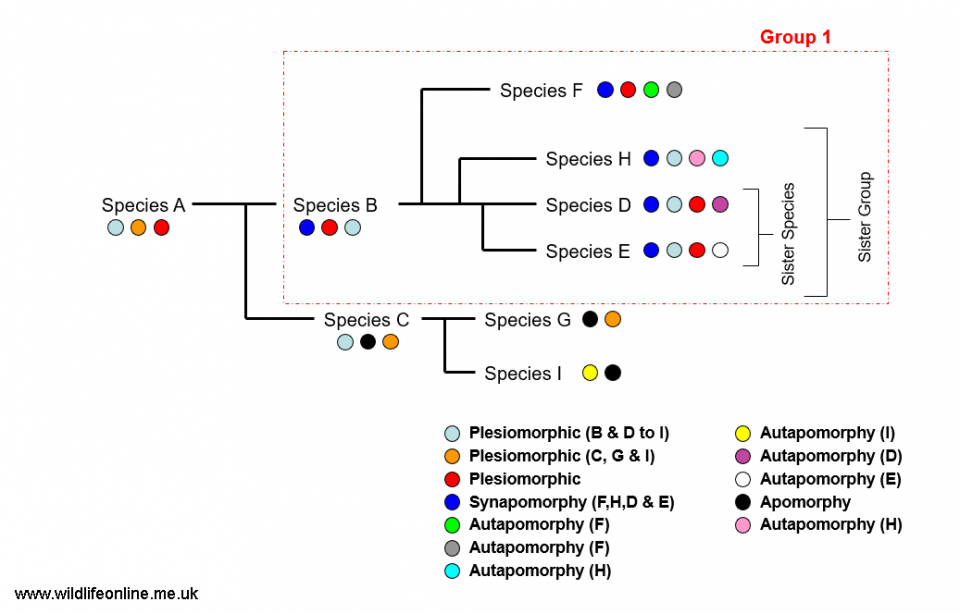
So, in order to build our scheme, we need to identify the organisms in which novel characteristics first crop up; taxonomists call these “branching points”. We start with a group of species and some data (genetic, anatomical, even behavioural) that characterizes them; you choose your characters/features and then you ‘weigh’ them in terms of how important you think they are – this is perhaps the most contentious step in the process and taxonomists frequently disagree on which characters should be used and how important they are.
Finally, you organize your subjects into groups on the basis of how many synapomorphies they possess. The result is a graph, called a cladogram, that represents the distribution of the characters; from this we can start to establish possible evolutionary relationships. Ultimately, the more synapomorphies there are among two species or groups, the more recently they shared a common ancestor and thus the more closely related they are likely to be.
If you find all of these groupings and terms mind boggling, you're not alone. The important point here is that we can group species based on shared characteristics and that there is a difference between describing something and defining it. Although the terms may appear superficially similar, they are actually crucially different and it's the difference that underpins our cladistic grouping.
Returning to our peregrine falcon example, you might describe it as a medium-sized predatory bird with a mottled brown-to-grey back, a white belly flecked with brown, and a bright yellow base to its beak. While someone else might be able to identify a peregrine based on this, does it really define what a peregrine is? The answer is no; there are several raptors with similar body colouration, and bright yellow bill-bases. So, in order to define what makes a peregrine a peregrine, we have to think about those features unique to it – those that aren't shared by any other creature. Only then can we say that the bird is a peregrine and not, say, a hobby (Falco subbuteo).
Displaying taxonomic relationships graphically
Diagrams represent a convenient method of expressing relatedness. In the case of taxonomic relationships they generally take the form of either a cladogram or a phylogenetic tree. Often, these terms are used interchangeably, not least because they share the same basic appearance, but some taxonomists argue that they are completely different. Whether you consider cladograms types of trees or not, the main difference between the two is that a cladogram doesn’t make a statement about evolutionary pathways, while a tree does. Instead, all a cladogram shows is the distribution of your chosen characters.
Cladograms
A cladogram is a branched diagram that shows patterns of relatedness; they look similar to a family tree turned on its side and are read left to right. (A few publications display cladograms vertically, in which case they’re read bottom to top.) In the example below, A represents the common ancestor of B, C and D. If you group A, B, C and D together they form a monophyletic clade because the group contains all descendants (B, C and D) of a common ancestor (A). B and C share more synapomorphies than either species does with D, making them sister taxa (i.e. they are more closely related to each other than anything else). In terms of descriptive terminology for cladograms, the first line (connecting A to the main graph) is referred to as the trunk (of the tree) and each point where the line splits in two is called a node; the lines themselves are referred to as lineages.
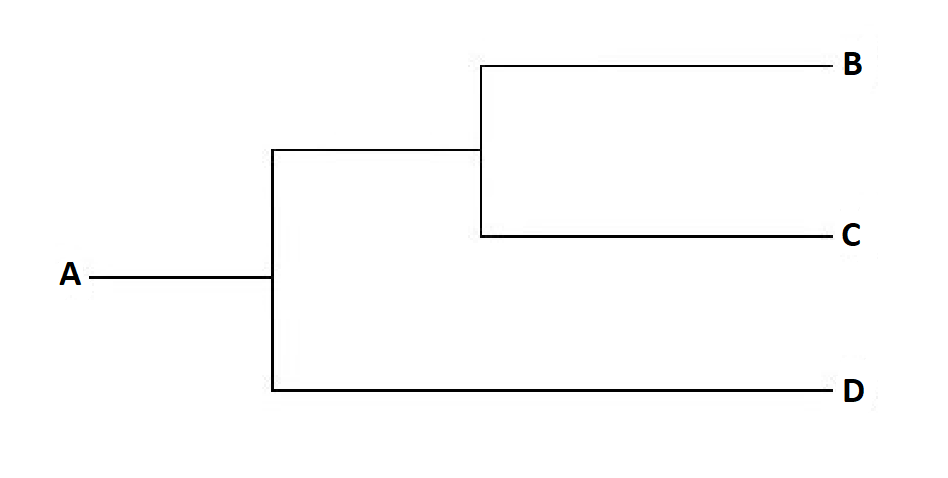
By this point, if you’re still with me, you may have noticed that if cladograms are created on the basis of the chosen characters and their weighting, then changing this ‘importance’ would result in a different graph being produced. Consequently, taxonomists divide (here we go again!) cladograms into two types. There are those that require only the minimum number of ‘steps’ (gains, losses or modifications of a character) necessary to explain the distribution of a character – these are the parsimonious or “optimal” cladograms.
Alternatively, there are those that require more steps – the “suboptimal” cladograms. In essence, the most parsimonious cladogram is the simplest, making the fewest assumptions and having the fewest steps in it. The potential for different characters and weighting to alter the end result, however, means that the most parsimonious graph is not necessarily always the best choice. Ultimately, only when several analyses using different sets of data point in the same direction can you be relatively sure that any resulting tree paints an accurate picture of the evolution of a group or species.
Phylogenetic Trees
Phylogenetic trees are branching diagrams, possibly a type of cladogram depending who you ask, that represent possible evolutionary pathways. The trees have branches, the length of which is proportional to the predicted (or hypothesised) time between the divergence of the organisms, groups or gene sequences.
The example above shows a cladogram (left) and one of 12 possible phylogenetic trees that can be generated based on it. The cladogram shows that the lizard and salmon share more inherited traits (synapomorphies) than either does with the shark or lamprey. In other words, as a group, the lizard and salmon have more in common with the shark than they do with the lamprey. The tree suggests that a hypothetical ancestor (Z) gave rise to the lamprey and to the shark; the scheme then goes on to imply that a hypothetical descendent of the shark (X) gave rise to the salmon and the lizard. The bar down the left-hand side of the tree signifies the time scale over which this is hypothesised to have happened, and is usually based on molecular data.
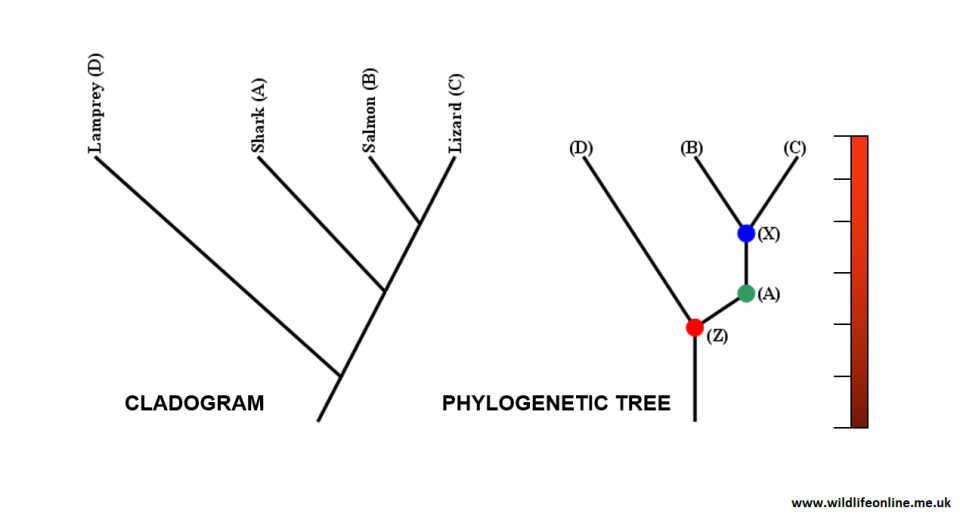
The origin of species
Following our trees to the end, their so-called “terminal taxa”, leaves us with that which we call a species; but what is a species, exactly? This is perhaps one of the most contentious questions in taxonomy. You’ve probably heard the term species used with an air of certainty, but we still don’t have an infallible definition of what makes something a species. The problem lies largely in our attempt to, as Charles Darwin put it, “define the indefinable”. The processes of evolution and speciation (the formation of new species) are continuous ones, which makes it difficult to group the results. This explains why there are currently so many proposed definitions (concepts) of what a species is. Perhaps the most well-known definition is the Biological Species Concept (BSC).
The biological species concept was first conceived by Ernst Mayr and proposes that species are "groups of actually or potentially interbreeding natural populations, which are reproductively isolated from other such groups". To put it another way, under the BSC a species is a group of individuals that freely interbreed with each other under ‘natural conditions’ (another sticking point) to produce offspring that can themselves breed. Some argue that this definition is weakened by animals such as ligers and tigrons (lion and tiger hybrids), but various parts of Mayr’s concept serve to exclude unusual or artificial matings, such as those resulting from human interference or those that may physically be able to interbreed but that don’t normally do so in the wild because they, for example, don’t live in the same country or inhabit different habitats such that they rarely or never come into contact.
There are, nonetheless, some issues with this theory. How do we know two species can’t interbreed unless we try mating them? Also, if we just consider geographic isolation, does this mean the red deer in the New Forest are a different species to those in France, given there’s the English Channel separating them and virtually no chance of them interbreeding without human intervention? Probably the biggest problem with the BSC is what to do with animals like sponges, planarians and echinoderms that don’t reproduce sexually. Despite these issues, it is fair to say that the BSC works well for most animals.
In a bid to address some of the gaps in the BSC, many other species concepts have been proposed and are currently about 26 different ones. Each concept tries to provide an all-inclusive definition of what it means to be a species, but none are without their problems. In terms of practicality, some biologists lean towards the General Lineage Concept (GLC). The GLC states that as different lineages evolve and diverge their genotype (genetic make-up) and phenotype (physical appearance) change to the point where, eventually, you can assign an animal to one species or the other. So, in essence the GLC and BSC aren’t all that different. The GLC is saying that species are lineages that retain their genetic integrity with respect to other lineages over time and space – i.e. they don’t merge/interbreed with each other.

The advent of molecular and genetic techniques has greatly enhanced our ability to assess what constitutes a species and taxonomic relationships. Molecular and genetic typing has seen to it that we are no longer restricted to basing our interpretations simply on how an organism looks and perhaps the biggest ‘rival’ to the BSC is now the Phylogenetic Species Concept (PSC), which does away with sex altogether.
The PSC centres on monophyly, stating that related organisms share characteristics because they share a common ancestor. You start with large groups and, based on synapomorphies, split them up into ever smaller ones until you arrive at a group that can be split no further: according to the PSC, this is a species. Some critics argue that the PSC leads to an ‘over splitting’ of species, although as Carl Zimmer pointed out in his article, many think we should just go where the data lead us rather than worrying about the number of species we end up with. Still, given that modern genetic analysis allows us to identify down to the individual, the potential for this ‘taxonomic inflation’ is considerable.
In the end, when trying to decide if the critter you’re looking at is a species in its own right, the best option is invariably to consider all lines of evidence at your disposal, and to include genetic data whenever possible. The jury is still very much out on the best way to proceed when it comes to defining a species, but the molecular and genetic tools at our disposal will no doubt play an increasingly large roll in future hypotheses.
Moving the goalposts
Those who do their best to follow the rather tumultuous world of taxonomy often become confused and frustrated when species are re-classified, especially if this happens several times in a short period. A good example of this is the taxonomic history of the sandtiger shark (Carcharias taurus), which Aidan Martin reviewed in an article on his site. The point to remember is that organisms aren’t re-classified capriciously or whimsically – any reassignments come about as a result of new evidence.
Hopefully, as Science forges ahead it will allow taxonomists to get a better handle on the interrelationships of the organisms on Earth and changes, while almost inevitable, will occur less frequently. In the meantime, as Aidan put it:
“Nature is messy; Science is tentative; as long as these truths remain relevant to biological research, scientific names will continue to be revised."